2007/10/26
2007/07/04
ooo
Labels: abiogenesis, bacteriology, biochemistry, biopoiesis, Cell Biology, evolution, Genetics, Geology, Immunology, intelligent design, mechanisms, medical science, philosophy, phylogeny, SET, virus
2006/12/31
overview
Proteins are essential to the structure and biological viability of all living cells and viruses. The cellular proteome is the total cellular protein under a particular set of conditions, while the complete proteome is the sum of all potential proteomes of a cell. Proteomics has become the subject of much research in cell and molecular biology.
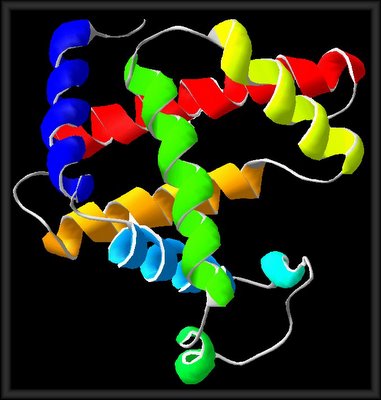
myoglobin, mod. gjh.md
Proteins play a number of vital roles as:
Enzymes or subunits of enzymes -- catalyzing cellular reactions.
Structural or mechanical roles -- structural components of tissues, components of the cytoskeleton, centrioles, cilia and flagella, microtubules, molecular motors.
Intra- and intercellular signalling functions -- ion channels, receptors, membrane pumps.
Regulatory proteins in genetic transcription, RNA processing, spliceosomes.
Products of immune response that aid in targetting of foreign substances and organisms
Storage and transport of various ligands.
The source of essential amino acids
Almost all natural proteins are encoded by DNA, which is transcribed and processed to yield mRNA, which then serves as a template for translation by ribosomes at the rough endoplasmic reticulum.
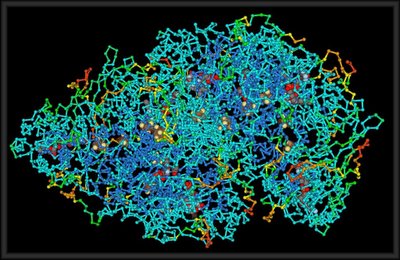
Structure of a protein, public domain
2006/12/30
apoptosis: Bcl-2 proteins
2006/12/29
BER glycosylases
2006/12/28
caspases
At least 14 caspase isoforms have now been identified. These isoforms are broadly categorised into initiators, effectors and inflammatory caspases. Initiator caspases such as caspase-8 and 9 cleave and activate effector caspases such as caspase-3. Effector caspases cleave specific proteins that ultimately leads to cell death by apoptosis. Caspase activity leads to a proteolytic cascade in which one caspase can activate other caspases. This amplifies the apoptotic signalling pathway.
Cleavage of proteins by caspases can either activate them (e.g. other caspases and ICAD), or they can deactivate them (e.g. PKB/Akt, Raf-1 and PARP-1). Generally, proteins that promote apoptosis are activated, and proteins that promote survival are inactivated." MORE
2006/12/27
DNA repair enzyme : hOGG1
DNA damage: eukaryotic response: NKG2D: RNR
MRC Radiation and Genome Stability Unit: "Radiation-induced damage in DNA has been shown to migrate and localise at guanine "
dNTP : "In eukaryotes, DNA damage elicits a multifaceted response that includes cell cycle arrest, transcriptional activation of DNA repair genes, and, in multicellular organisms, apoptosis. We demonstrate that in Saccharomyces cerevisiae, DNA damage leads to a 6- to 8-fold increase in dNTP levels. This increase is conferred by an unusual, relaxed dATP feedback inhibition of ribonucleotide reductase (RNR). Complete elimination of dATP feedback inhibition by mutation of the allosteric activity site in RNR results in 1.6-2 times higher dNTP pools under normal growth conditions, and the pools increase an additional 11- to 17-fold during DNA damage. The increase in dNTP pools dramatically improves survival following DNA damage, but at the same time leads to higher mutation rates. We propose that increased survival and mutation rates result from more efficient translesion DNA synthesis at elevated dNTP concentrations."
Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L.
Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase.
Cell. 2003 Feb 7;112(3):391-401.
Comment on: Cell. 2003 Feb 7;112(3):286-7.

